The GPCR Assay Service Center at Penn State College of Medicine was established to accelerate scientific discovery in G protein-coupled receptors (GPCRs) physiology and pharmacology.
It is estimated that approximately 25% of all FDA-approved drugs target GPCRs. The GPCR core provides investigators the ability to begin to identify signal transduction processes that are specific to their GPCR of interest (such as opioid, cannabinoid and muscarinic) that is either natively present or expressed heterologously in cell systems.
Signal transduction following GPCR stimulation is typically monitored by measuring changes in cell membrane potential or ions (i.e., Ca2+, Na+, K+) employing fluorescent dyes. The core will assist investigators with drug development studies. The core will support researchers with assay development and optimization, pilot data, and grant assistance. The GPCR core houses all the necessary instrumentation used.
Jump to topic
Search
Instrumentation and Services
Equipment
- FlexStation 3 Multi-Mode Microplate Reader (Molecular Devices)
- Neon NxT Electroporation System (Thermo-Fisher)
- Countess 3 Automated Cell Counter (Thermo-Fisher)
Services
- Identifying GPCR signal transduction pathways
- GPCR ligand concentration-response relationships
- GPCR pharmacological profiles
- Heterologous expression of GPCR of interest
- Data analysis
Example of results
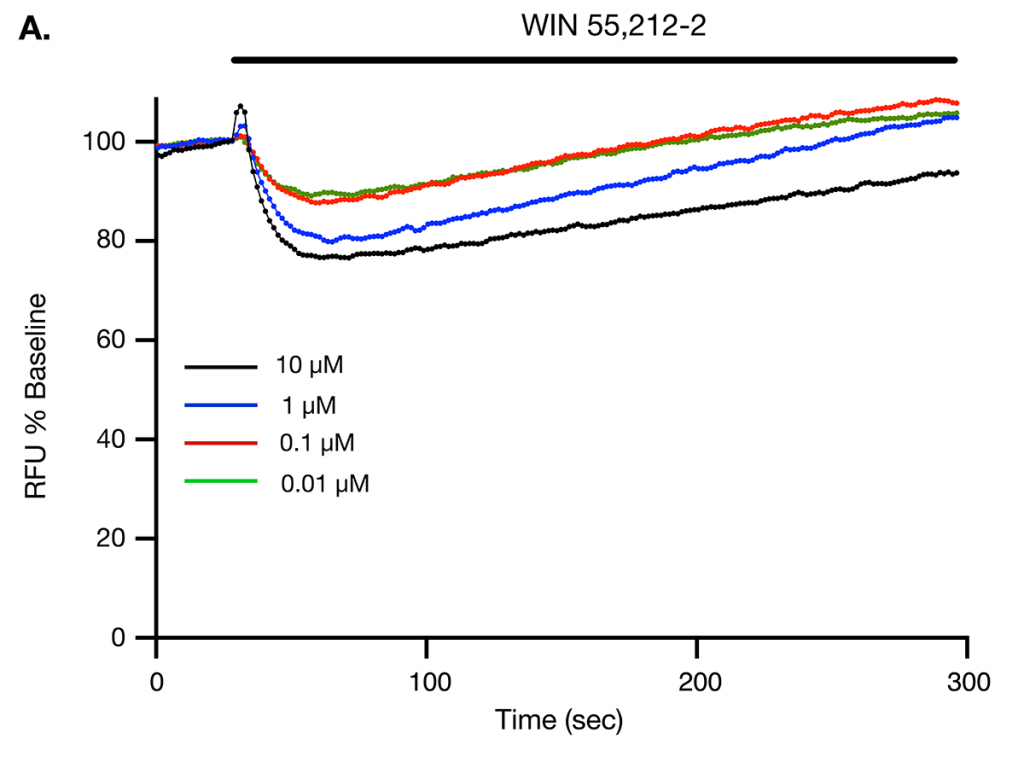
WIN 55,212-2 causes membrane hyperpolarization in CHO cells transfected with CB1 and GIRK channel cDNA.
Figure A: Raw traces show decrease in fluorescent signal following application of 10, 1, 0.1 and 0.01 μM WIN 55,212-2 to CHO cells expressing human CB1 receptor and G protein-inwardly rectifying K+ (GIRK) channels. The decrease in fluorescence indicates membrane hyperpolarization from GIRK channel activation. The Y-axis indicates the raw fluorescent units (RFU). The agonists were added for the duration of the bar.
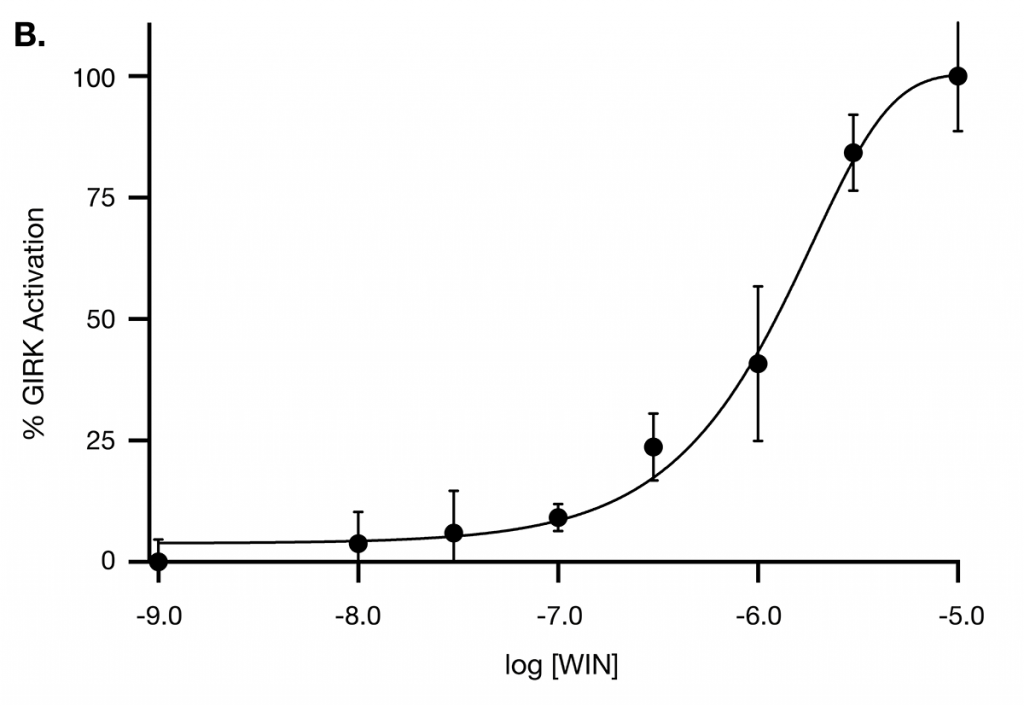
Figure B: Concentration-response relationship of the WIN 55,212-2-mediated GIRK channel activation. Each data point represents the mean (±SD, n=4 to 8 wells). The smooth curve was obtained by using the least squares regression and fitting the points to the Hill equation.
Procedures, Protocols and Forms
All publications, press releases or other documents that result from the utilization of any Penn State College of Medicine Institutional Research Resources including funding, tools, services, or support are required to credit the core facility and associated RRID.
Working with the GPCR core
For scheduling and details on fees, contact Dr. Ruiz-Velasco, director, at vjr10@psu.edu or 717-531-6076.