Use the buttons below to learn about Dr. Parent and her team.
About the lab
We are interested in understanding how retroviruses produce new virus particles that are released from cells to spread infection. Our focus is on the retroviruses HIV-1 (human immunodeficiency virus type 1), the cause of AIDS, and RSV (Rous sarcoma virus), the first virus discovered to cause cancer. Our long-term goal is to identify potential therapeutic targets by uncovering how retroviruses hijack host cell machinery, with a focus on nuclear factors. We have recently begun biophysical studies of ribonucleoprotein complexes that undergo biomolecular condensate formation. We also study SARS-CoV-2, the virus that caused the COVID-19 pandemic, with a focus on developing strategies to interfere with virus production. Our research program is currently funded by National Institutes of Health R01 and R21 grants.
Jump to topic
Search
Meet the Team

Rebecca Kaddis Maldonado, PhD
Faculty Researcher
Dr. Maldonado is an expert in all aspects of virology research with specialization in advanced fluorescence microscopy including live-cell imaging, particle tracking and super-resolution imaging.

Greg Lambert, PhD
Postdoctoral Scholar
Dr. Lambert is focusing on the role of biomolecular condensates in retroviral RNA packaging and virus assembly.

Jordan Chang, MS
Graduate Student
Jordan’s primary work is on HIV-1 Gag interactions with chromatin and transcription machinery.

Malgorzata (Gosia) Sudol, MS
Senior Technician
Malgorzata performs molecular biology experiments and protein purifications to support the work of everyone in the laboratory.
Research Topics and Current Projects

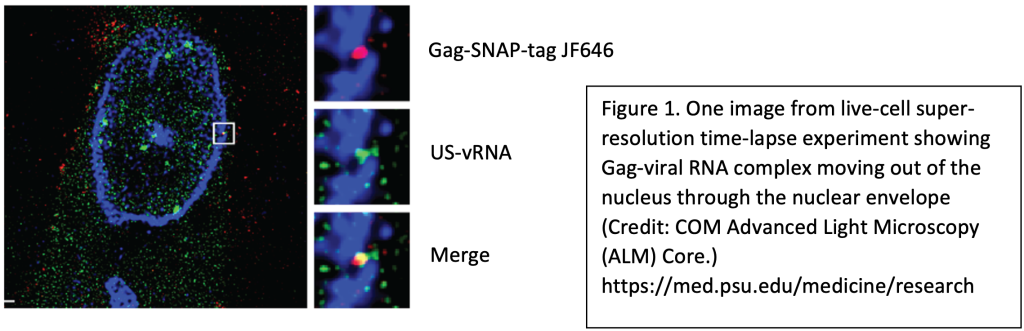
Using single molecule detection methods and live-cell confocal imaging with super-resolution stimulated emission depletion (STED), we have visualized dynamic trafficking of RSV viral complexes. Our data suggest that RSV proteins capture viral RNA at the proviral transcriptional burst site in the nucleus. We are testing the hypothesis that retroviral RNAs are sorted co-transcriptionally to determine their fates as mRNA or genomic RNA, which is packaged into assembling virions released from the plasma membrane.
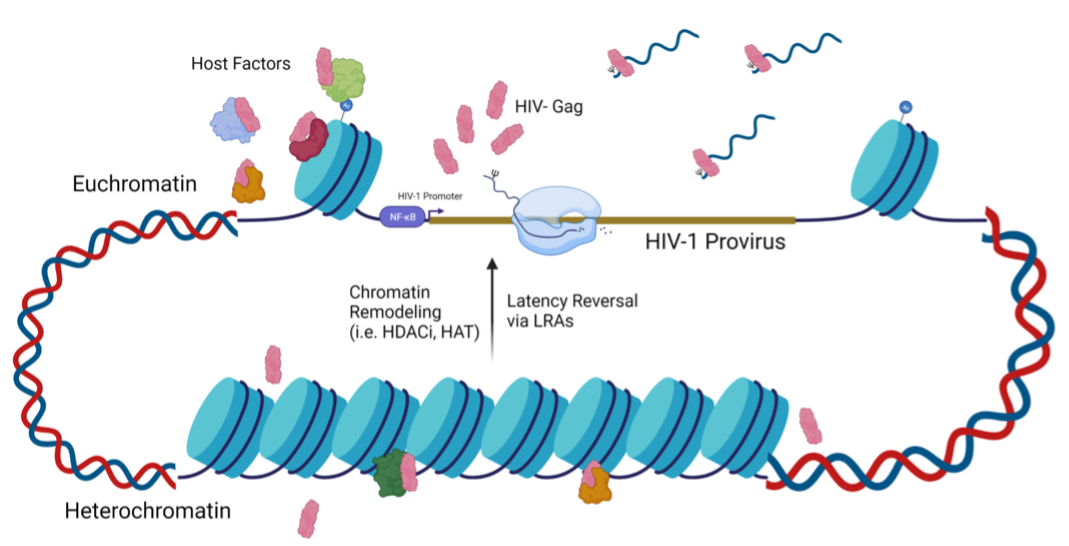
This project aims to characterize the spatiotemporal properties of HIV-1 Gag by examining how and where Gag localizes to the nucleus. Our studies indicate that Gag preferentiallylocalizes to the euchromatin, where active transcription occurs. We hypothesize that Gag localizes to active viral transcription sites to select for genomic RNA to package into new virions. Currently, we are investigating Gag’s hijacking of transcription machinery to facilitate retrovirus replication.
Retroviral Gag proteins perform the critical function of selecting and packaging two copies of unspliced viral RNA to serve as the genome for each nascent virion. Our lab discovered that RSV Gag traffics to the nucleus and colocalizes with nascent viral RNA at sites of transcription. We identified several components of RNA polymerase II-associated transcription machinery that associate with Gag proteins, supporting the novel hypothesis that RSV Gag hijacks transcription machinery to facilitate genomic RNA selection.
Contact Us
 Dr. Leslie J. Parent
Dr. Leslie J. Parent
Vice Dean for Research and Graduate Studies
Email: lparent@pennstatehealth.psu.edu
